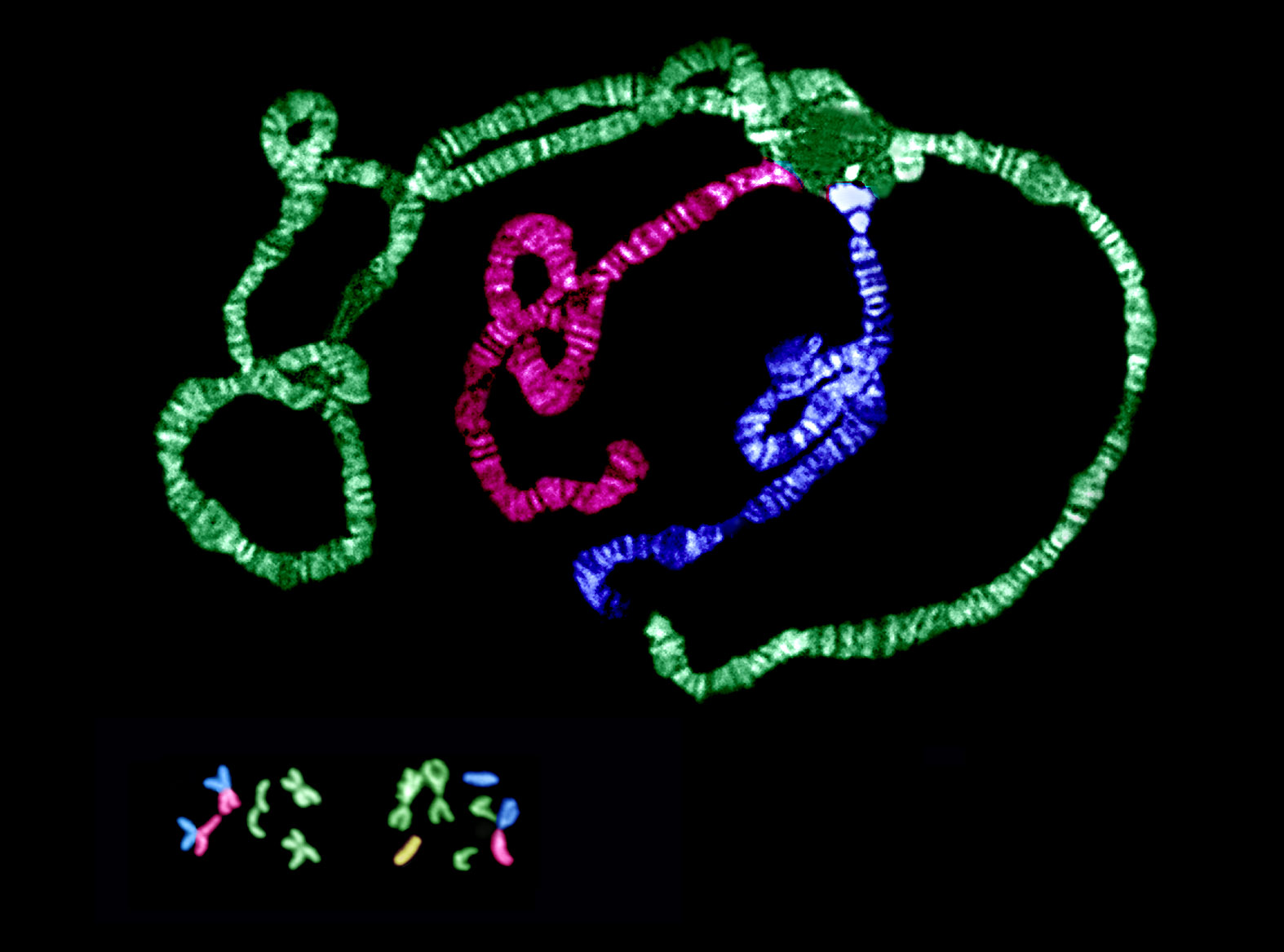Bryant McAllister
Research summary
Evolutionary genetics and genome evolution
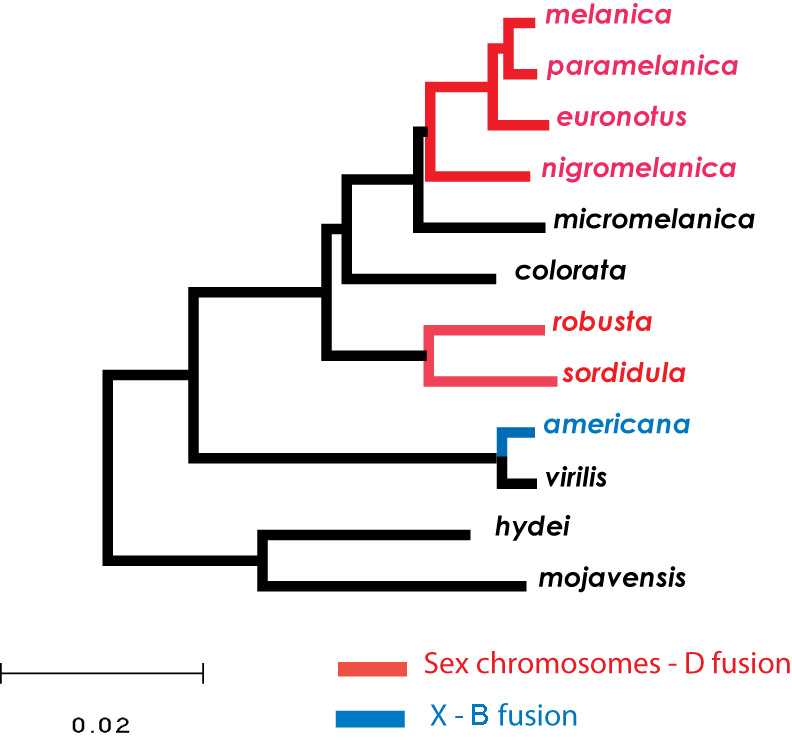
My research interests are in the field of evolutionary genetics, especially understanding processes occurring through or influenced by genome-level mechanisms. Active research projects in the lab use flies in the genus Drosophila for empirical studies combining molecular, cytological, and genetic approaches. Three primary research areas are currently being pursued in the lab: 1) identifying the role of chromosomal rearrangements in facilitating adaptive evolution, 2) determining the consequences of sex-linked transmission, and 3) defining common pathways of sex-chromosome evolution.
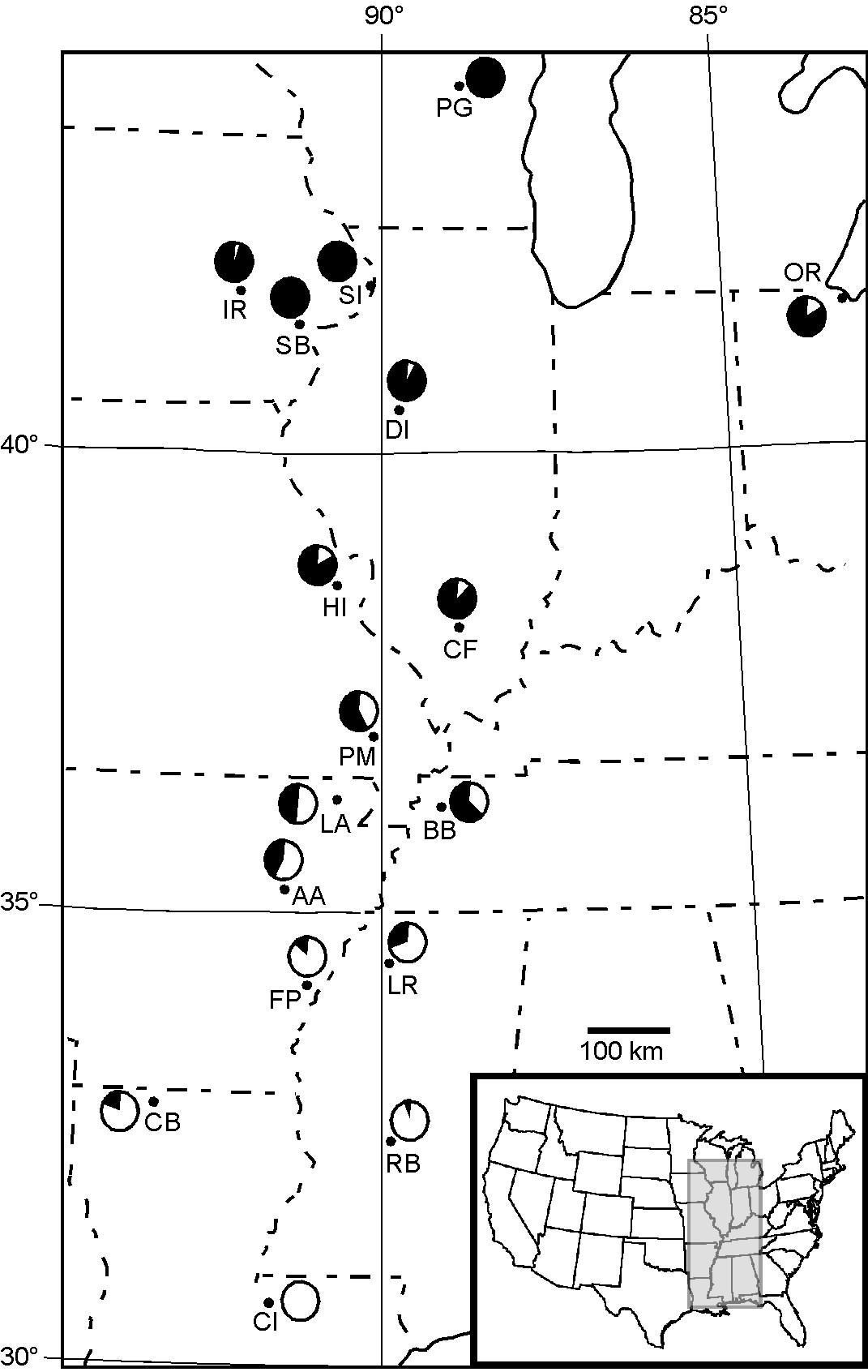
Changes in chromosomal arrangement are common, but their significance is unknown. We are currently examining how natural selection influences a chromosomal rearrangement involving a centromeric fusion of the X chromosome and an autosome in Drosophila americana (see figure). This derived arrangement coexists with the ancestral arrangement, and their frequencies exhibit a strong latitudinal cline in the central and eastern US. Through integrated studies of chromosomal and nucleotide variation, we are examining a long-standing hypothesis that modification of crossing over between alternative chromosomal arrangements coordinates adaptive genetic variants.
Our projects on sex chromosomes are focused on understanding both primary and secondary mechanisms of change during the emergence of this unique chromosomal pair following their acquisition of the primary sex-determining locus. This transition from autosomal pair to sex-chromosome pair is well documented—mostly using plant and vertebrate systems, including humans. One line of research capitalizes on the very recent occurrence of the X-Autosome fusion in D. americana to examine the initial causes of divergence between a chromosomal pair experiencing sex-linked transmission. Current results demonstrate that divergence is driven by selection, and quite surprisingly, optimization of female fitness at a cost to male fitness may be the underlying mechanism.
We are also investigating other Drosophila species with similar rearrangements involving the sex chromosomes. These comparative analyses provide a framework for identifying common features in the transition from autosome to sex chromosome, such as determining how genes on the Y chromosome are silenced, defining the relationship between functional degeneration on the Y and dosage compensation on the X, and estimating the temporal dynamics of this transition.
Keywords: Sex chromosome evolution, maintenance of chromosomal variation, and molecular population genetics using Drosophila as a model
Selected images