Robert Malone
Research summary
Chromosome behavior in meiosis
Chromosomes undergo several special events during meiosis, the process wherein chromosome number is reduced from diploid to haploid ( to form sperm, pollen, eggs, spores, etc.). The first unique chromosomal event is genetic recombination and homologous chromosome pairing. The second unique event, the reductional division, segregates the two paired homologous chromosomes to opposite poles. We are answering three major questions about chromosomes in meiosis utilizing genetics, molecular biology, microscopy, and protein biochemistry.
First, we are asking how meiotic recombination initiates. Specifically, we are asking how the 10-12 gene products required interact with each other to form an initiation complex; in concept this complex is analogous to the complexes that initiate DNA replication. We have begun to define interactions between proteins that are only made in meiosis, and whose only role is to initiate recombination by making breaks.
Second, in collaboration with John Logsdon's group, we are asking if meiotic functions involved in recombination and the reductional division present in other organisms have conserved functions. Logsdon's analysis strongly suggests that meiosis evolved very early during the evolution of eukaryotes, perhaps simultaneously. The conclusion would be strengthened by finding that genes in other eukaryotes had retained function. We have devised expression vectors and assay systems to detect meiotic function of conserved genes that are sensitive over a 200 fold range.
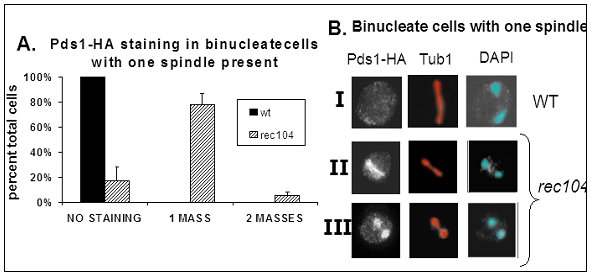
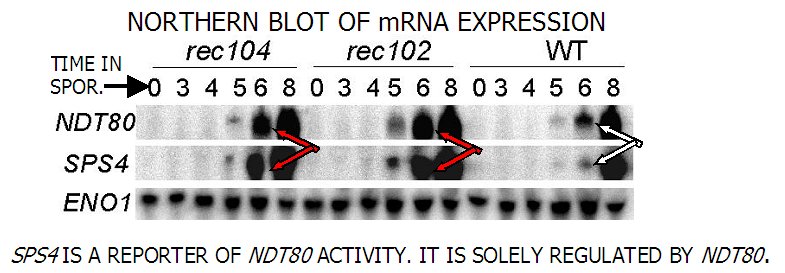
Third, we have discovered that the initiation of recombination somehow signals the reductional division so that it delays until the proper time. This communication is an intracellular signal transduction process. We have determined that 8 of the 10 initiation genes are required for this signal. In addition, 2 genes involved in the special meiotic recombination structure, the synaptonemal complex, are also required for the normal delay. We are now asking how the cell detects the initiation signal, if their assembly on the chromatin is the signal, and how the signal is transduced to create the proper delay of the division. We have identified one of the targets of the signal, a major transcriptional regulator of middle meiotic genes. We are also looking for mutants that cannot carry out the proper communication between recombination initiation and the first division, in order to define the components involved in the event.
Selected images